GFF3 Tab
If you have a previous annotation, or results from other annotation tools, in the GFF3 format, the results can be uploaded to GenSAS and used during the annotation process. The GFF3 format is a standard 9-column file that has specific information in each column. Basically a GFF3 file lists the locations of features (genes, alignments, repeats) on a specific DNA sequence. GFF3 files must have been created from the sequence you uploaded into GenSAS. For more details about the GFF3 format, please see the GMOD wiki. For GenSAS, it is important to make sure that the sequence names in column 1 of the imported GFF3 file, exactly match the sequence name being used in GenSAS (Fig. 12). Please see the information about sequence names in GenSAS on the Sequences Tab page in this guide. If the sequence names in column 1 of the GFF3 file do not match the sequence names in GenSAS, then the data will not be imported into GenSAS.
.jpg)
Figure 12. Example of GFF3 file format.
There are five different GFF3 file import options; Repeats, Transcript Alignments, Protein Alignments, Gene Predictions, and Other Features (Fig. 13). Each importer will recognize and import certain feature types in the GFF3 file (Table 1). Feature types are listed in column 3 of the GFF3 file. All imported GFF3 data can be viewed in JBrowse, but some of the data can also be used during the annotation process. For example, the repeat data will be available under the Masking step and the transcript data will be available for use to make a gene model consensus. The "GFF3 Files" option (Fig. 13A) will display a list of all GFF3 files that have been uploaded by the user and whether they are use in the current project or other projects. Files that are not in use, can be deleted by the user.
| GFF3 Importer | Recognized feature types |
|---|---|
| Repeats | repeat, repeat_region |
| Transcript Alignments | match, match_part |
| Protein Alignments | match, match_part |
| Gene Predictions | gene (required), transcript, mRNA, CDS, exon, five_prime_UTR, three_prime_UTR |
| Other Features | any term in column 3 |
Table 1. GFF3 feature types recognized by the GFF3 importers in GenSAS.
To import a GFF3 file, click on the appropriate importer for the data type you are uploading. Use the "Choose File" option to select a new GFF3 file and then click "Upload"(Fig. 13B). If the GFF3 file has been previously loaded into GenSAS, use the pull-down menu under "GFF3 File" (Fig. 13C) to select the file. Type in a Job Name and click "Import File" and a job will appear in the Job Queue. If you have different GFF3 files to load under the same importer type, you can set-up multiple jobs. Each job uploads a single file, and each job must have an unique name. Please make sure your job names are meaningful so you know what data was imported by that job when viewing the data later in GenSAS.
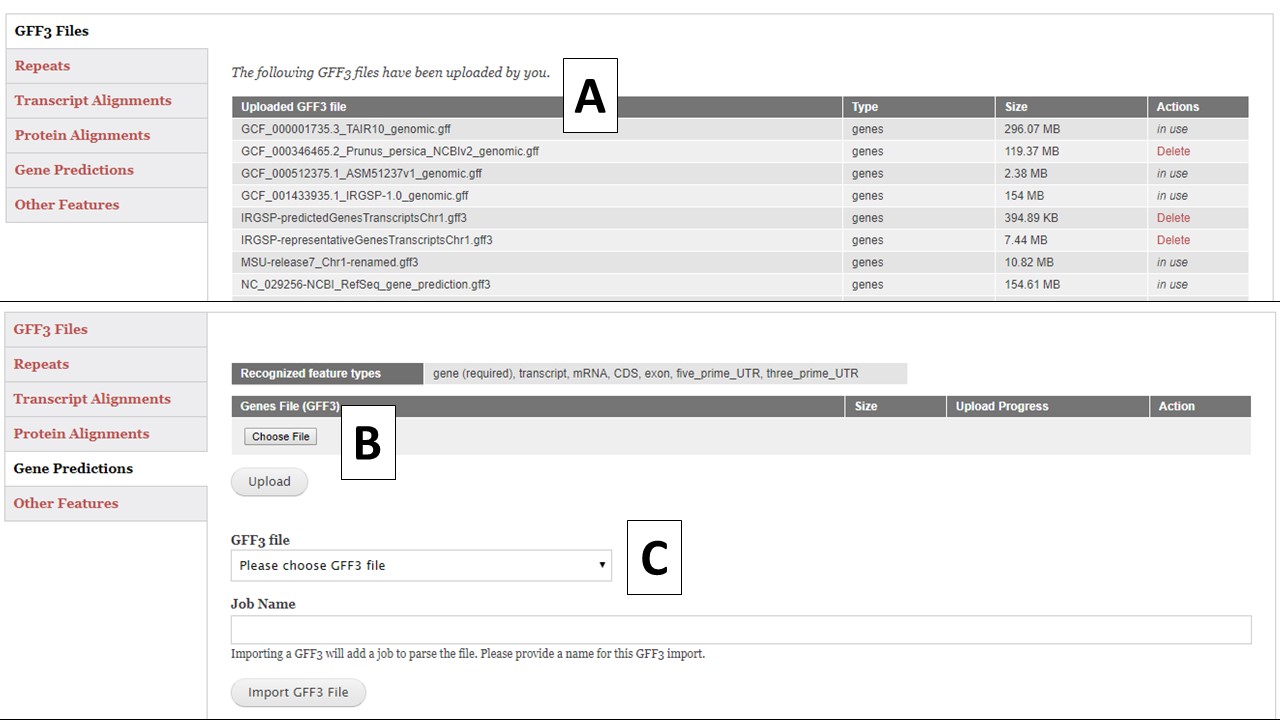
Figure 13. GFF3 tab in GenSAS.
Once you have submitted the import jobs, GenSAS will process the files and once the jobs are complete, you will be able to open JBrowse and view the data. Open JBrowse by clicking on the "Browser" link in the right hand menu and then click "Open Apollo". When you are done uploading GFF3 files, click "Proceed to next step", which is located near the top of the tab, to move on to the next step of the annotation process.
If your GFF file is a eukaryote based genome project which also includes chloroplast, mitochondria, plastid, or plasmid DNA as scaffolds please be aware that the gene models for those sequences will not import into JBrowse. For eukaryote projects, GenSAS displays mRNA features in JBrowse/Apollo. These sequences are prokaryotic in origin and usually do not have mRNA features in the annotations.




